Introduction to the PC-NPGRIS
Data Management
System of Chinese Taipei
Shu Chen and Ming-Jen Fan
Taiwan Agricultural Research
Institute-NPGRC
Wufeng, Taichung, Taiwan
41301
npgris@hgp40.npgrc.tari.gov.tw
Abstract
The PC-NPGRIS
Data Management System was designed for breeders and their allied workers
within our Economy to input, verify, and transfer their data to the NPGRIS main
database server easily. The
information also can be managed as a mini-database owned by the agricultural
workers themselves. Now, we offer
this idea to APEC Economy Members.
The PC-NPGRIS have been modified to include an English version in order
to provide a simple and useful tools for other Members to establish a
mini-database on germplasm owned by themselves. The hardware and database architecture will be described in
detail.
Introduction
Plant germplasm
is the main source of developing new crop cultivars and the basis of crop
improvement. It is well recognized
that an efficient data management system of germplasm can help breeders and
related scientists to search for needed germplasm information and select
appropriate materials to be utilized in crop improvement.
Our National
Plant Genetic Resources Information System (NPGRIS) was established in 1993. There are three main activities of the NPGRIS.
(1) to set up a
database for all crop cultivars in the country, including related germplasm
information for breeders and allied researchers to search for needed
information.
(2) to develop a
genetic resources information network within our country and hopefully around
the world to strengthen the communication of information on germplasm.
(3) to handle the
processing and storage activities of germplasm at the National Plant Genetic
Resources Center (NPGRC).
In order to
accelerate the exchange of plant germplasm information among breeders and related
scientists all over the world, the NPGRIS has also upgraded its hardware and
entered the World Wide Web since 1997.
There are multi-functions in our web including the introduction to
NPGRC, database query, plant germplasm catalogs, plant genetic resources
newsletter, etc. (http://www.npgrc.tari.gov.tw or http://192.192.196.1)
Currently, there
are about 30,000 records of passport data, 20,000 records of characterization
data and 1,000 records of image data had been built in the NPGRIS database (Table
1). These accessions represent
more than 119 families, 367 genera and 500 species of plant germplasm. Most of the data were provided by the
breeders and related evaluators.
To ensure the accuracy of the data, we designed the end-user application
program for the scientists to input, verify, transfer their data to NPGRIS
easily, and manage the mini-database by the workers themselves. This end-user application program was
called “PC-NPGRIS Data Management System”.
The main
objective of this paper is to introduce the PC-NPGRIS Data Management System. The hardware and database architecture regarding how
PC-NPGRIS Data Management System was linked to the NPGRIS main database server
are also described in this paper.
Now, we wish to
extend this PC-NPGRIS idea to APEC Economy Member. We have modified the programs and translated the
language into an English version
in order to provide a simple and useful tool so that other Members may
establish an mini-database on germplasm owned by themselves.
Hardware
Architecture
The frame of
hardware architecture of NPGRIS is illustrated in Figure 1. The ORACLE database
management system and UNIX operation system are built in the HP9000 800/G40
database server. To modernize and
computerize the operations of the NPGRC, all the personal computers of
NPGRC-related laboratories are capable of being linked to the database server
with a client-server local network architecture. For information exchange, the NPGRIS has been linked to
other genetic resources centers around the world through the World Wide Web.
In order to
strengthen the communication of germplasm information within our Economy, the
NPGRIS provides telephone dial-up system for agricultural scientists of other
29 research institutions within our Economy to access information. Besides, interested researchers can also enter into the NPGRIS
database through network within our Economy such as TANET, Hinet or SeedNet. Thus, they can not only search for
needed germplasm information, but also send data from PC-NPGRIS Data Management
System to NPGRIS database server.
Database
Architecture
The NPGRIS
employs ORACLE as a relational database management system. The NPGRIS database has stored the
related data of plant germplasm, including passport data, characterization data
and image data. The passport data
and characterization data are both usually provided by the breeders or
collectors. To ensure the accuracy
of the data, the “PC-NPGRIS Data Management System” was developed for the
breeders to input, verify, transfer their data to NPGRIS, and manage as a
mini-database owned by the breeders themselves.
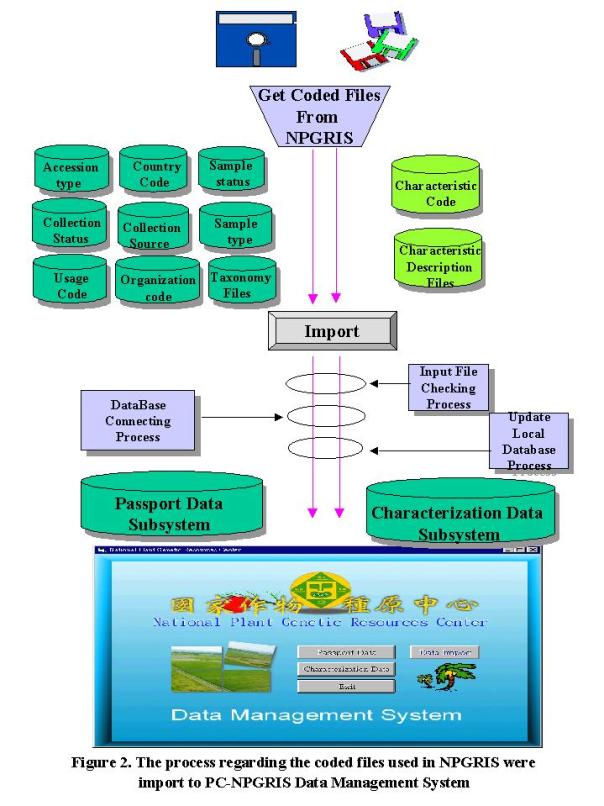
Because the
PC-NPGRIS Data Management System was based on the Microsoft Window operation
system, its operation is very easy and user-friendly. In addition, all of the coded files used in NPGRIS database
server must be import to PC-NPGRIS Data Management System in order to ensure
the consistent data format (Figure 2).
These coded files including accession type, country code, organization
code, sample status, collection status, collection source, sample type, usage
code and taxonomy files were entered in the PC-NPGRIS passport data
subsystem. The plant
characteristic code and its description files were used in PC-NPGRIS characteristic
data subsystem. In such a way, the
users just need to pop-down the code list, select and fill data in PC-NPGRIS
directly. The key-in operation
will be reduced¾, this is the
another benefit of PC-NPGRIS.
The passport data
record the background information of germplasm, such as scientific name,
pedigree, donor, collector, location of collection, criteria of distribution,
etc. In our PC-NPGRIS passport
data subsystem, these data fields were divided into 6 display pages such as
basic information, scientific name, pedigree, cultural practice, collection
data and other notes. The users
can click the menu bar on the top of the passport data subsystem to choose the
display page he want to fill in (Figure 3A). We also design the option buttons on the right side of the
passport data subsystem for the users to add, update, delete, save, query and
find data records with a “previous-next” options conveniently. The option buttons can also help the
users to export data files to NPGRIS main database server or print these data
on printed document. The program
flow of PC-NPGRIS passport data subsystem is shown in Figure 4.
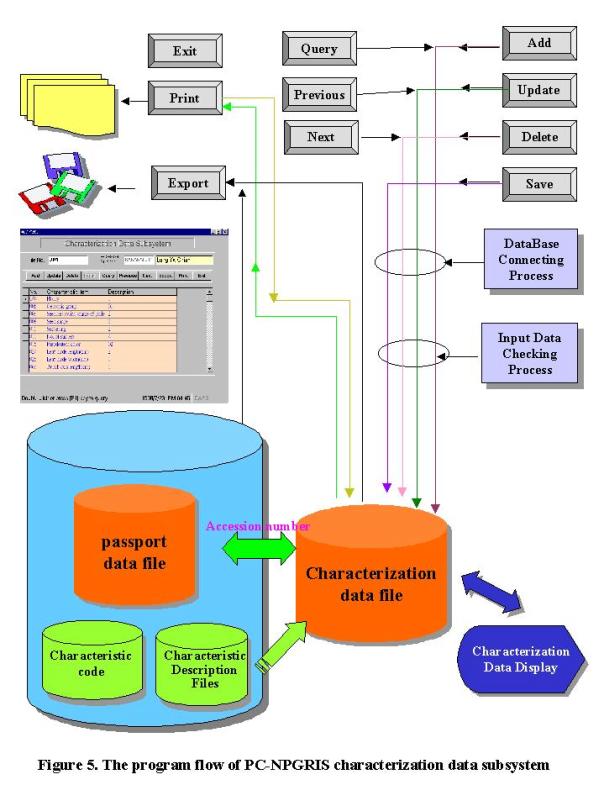
The
characterization data record the characteristics of a germplasm accession, such
as agronomic traits, resistance and tolerances to stress, chemical composition,
etc. As the NPGRC preserves
numerous plant germplasm accessions and the characteristic items of each
species are different, we organized crop committees to define the uniform list
of items and related descriptor states according to different crops. This uniform format of characteristic
descriptors of plant can also enable our researchers to follow. Currently, there are formats of 72
descriptors in NPGRIS (Table 2).
In the PC-NPGRIS characterization data subsystem, users can pop-down the
list of characteristic items to choose the described ones. Once the item is selected, the
description and its related code will be pop-down. The users can input data directly. These characterization data will be integrated with passport
data by accession number. We also
design the option buttons on the top of the characterization data subsystem for
users to add, update, delete, save, query and find data records with “previous-next”
option, as well as to export data files or print on paper (Figure 3B). The program flow of PC-NPGRIS
characterization data subsystem is shown in Figure 5.
Table
1. Statistics on the NPGRIS
|
Crop |
Scientific Name |
No. of Passport data |
No. of Characterization
data |
|
Soybean |
Glycine max |
12922 |
5986 |
|
Tomato |
Lycopersicon esculentum |
4339 |
0 |
|
Mungbean |
Vigna radiata |
4184 |
8330 |
|
Rice |
Oryza sativa |
4150 |
8659 |
|
Pepper |
Capsicum annuum |
2522 |
23 |
|
Common Flax |
Linum vsitatissimum |
1811 |
0 |
|
Sugarcane |
Saccharum officinarum |
1397 |
568 |
|
Peanut |
Arachis hypogaea |
1381 |
1349 |
|
Asparagus Bean |
Vigna nguiculata |
1020 |
219 |
|
Corn |
Zea mays |
746 |
193 |
|
Sweet Potato |
Ipomoea batatas |
483 |
0 |
|
Chinese
Cabbage |
Brassica campestris Pekinensis group |
470 |
0 |
|
Eggplant |
Solanum melongena
|
374 |
181 |
|
Sorghum |
Sorghum bicolor |
370 |
0 |
|
Medicinal Crops |
Medicinal Crops |
256 |
0 |
|
Peach |
Prunus persica |
230 |
157 |
|
Araceae |
Araceae |
203 |
0 |
|
Snap Bean |
Phaseolus vulgaris |
201 |
116 |
|
Jute |
Corchours capsularis |
188 |
0 |
|
Mango |
Mangifera indica |
174 |
22 |
|
Snow Pea |
Pisum sativum |
162 |
45 |
|
Adzuki Bean |
Phaseolus angularis |
155 |
150 |
|
Muskmelon |
Cucumis melo |
153 |
102 |
|
Edible Amaranth |
Amaranthus tricolor |
149 |
0 |
|
Morus |
Morus sp. |
209 |
0 |
|
Sunflower |
Helianthus annuus |
125 |
0 |
|
Taro |
Colocasia esculenta |
125 |
35 |
|
Bottle Gourd |
Lagenaria siceraria |
121 |
52 |
|
Watermelon |
Citrullus lanatus |
113 |
43 |
|
Asparagus |
Asparagus officinalis |
109 |
0 |
|
Cucumber |
Cucumis sativus |
104 |
48 |
|
Fruit Trees |
Fruit Trees |
101 |
0 |
|
Tea |
Camellia sinensis |
101 |
84 |
|
Banana |
Musa spp. |
100 |
63 |
|
Other < 100 accession |
|
2696 |
136 |
|
Total |
|
30927 |
20619 |
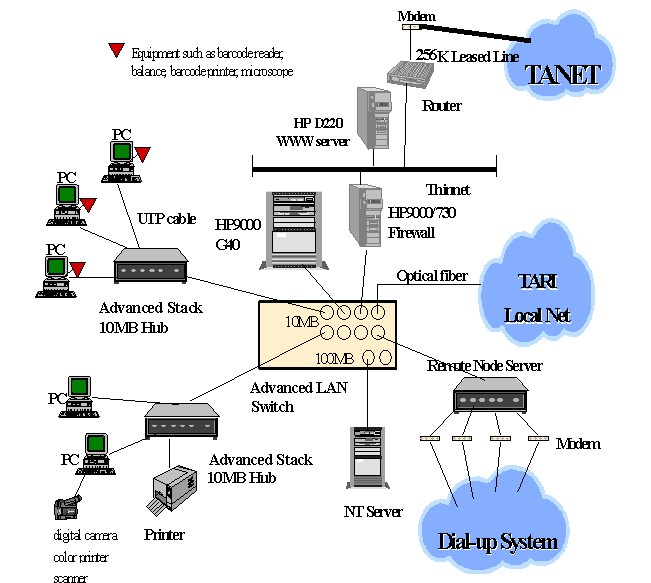
Figure 1. The
hardware architecture of NPGRIS
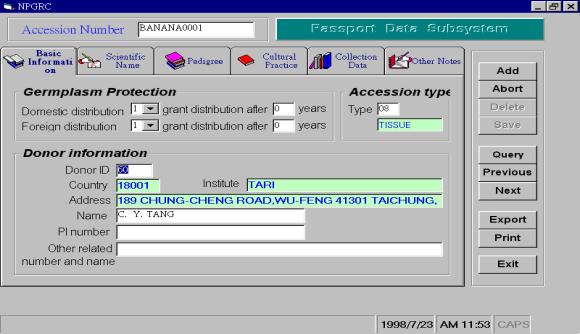 A
A
B
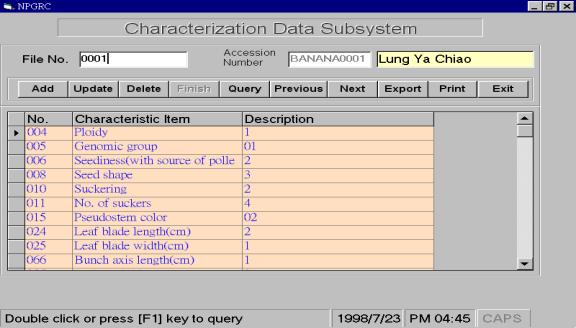
Figure 3. The English version of data management
system is developed in Microsoft window operation system.
(A) The windows of the
passport data subsystem on the screen
(B) The windows of the characterization data subsystem on the screen
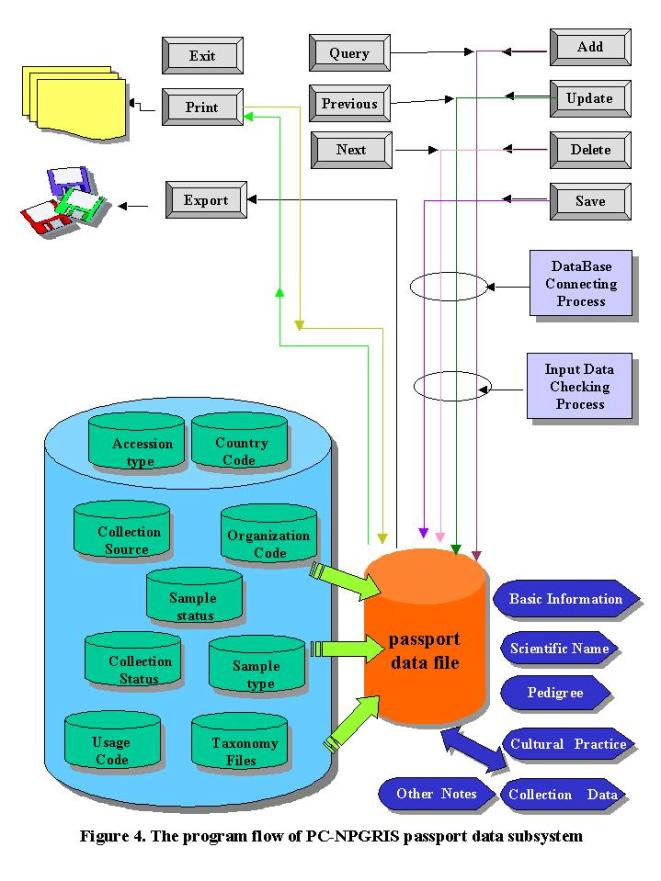
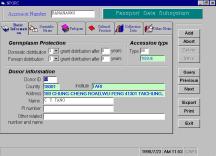
Table 2. Crops in the NPGRC with characteristic
descriptors format
|
Crops |
Number |
|
Food Crops: |
7 |
|
RICE、SOYBEAN、PEANUT、CORN、MUNGBEAN、ADZUKI BEAN、SWEET POTATO |
|
|
Vegetable: |
37 |
|
CABBAGE、MUSTARD、CHINESE CABBAGE、CAULIFLOWER、VEGETABLE SOYBEAN、SNOW PEA、KOHLRABI、CHINESE KALE、RAPE、BROCCOLI、BITTER MELON、CHINESE MUSTARD、RADISH、ASPARAGUS BEAN、LIMA BEAN、SNAP BEAN、WAXGOURD、WATERMELON、SQUASH、BOTTLE GOURD、POTATO、PEPPER、ORIENTAL PICKLING MELON、MUSKMELON、EGGPLANT、LUFFA、WATER CONVOLVULUS、CELERY、TOMATO、CUCUMBER、GARLIC、LETTUCE、EDIBLE AMARANTH、TARO、STRAWBERRY、ALLIUM SPP. 、ASPARAGUS |
|
|
Fruit Crop: |
18 |
|
LITCHI、LOGAN、BANANA、PAPAYA、GRAPE、MANGO、PERSINMMON、LOQUAT、PLUM、PEAR、PINEAPPLE、PEACH、APPLE、INDIAN JUJUBE、CITRUS、GUAVA、CARAMBOLA、WAX APPLE |
|
|
Special Crops: |
6 |
|
SUGARCANE、MULBERRY、TEA、HSIAN-TSAO、YAM、ANUECTOCHILUS |
|
|
Forage: |
2 |
|
GRAMINEAE FORAGE、LEGUMINOSAE FORAGE |
|
|
Other: |
2 |
|
CYMNOSPERMAE、ANGIOSPERMAE |
|
|
TOTAL |
72 |